Human Brain Project (HBP)
The Human Brain Project is a European-funded research initiative involving more than 100 institutions all over the continent. The goal of the project is to unveil the secrets of the human brain and uncover the mechanisms responsible for cognitive processes, learning and consciousness. The key-word for HBP is multi-disciplinarity: HBP is not only a fruitful ground for pioneering research in the field of neuroscience, but also a successful experiment of collaboration between several different scientists, bridging together medical doctors, biologists, physicists, computer scientists, engineers and philosophers, and linking experimental data with theoretical models and high-performance computer simulations.
HBP started in 2013, with the so-called ramp-up phase, during which the backbone and the main scientific targets of the project have been defined. In this starting phase, calls have been opened to the scientific community in order to extend the objectives of research and to address specific goals. A dedicated scientific board selected the projects that won the competition among the several applicants, forming what is now called the Systems and Cognitive Neuroscience Sub-Project (SP3). One of the 4 winner projects was WaveScalES, led by Pier Stanislao Paolucci. With it, APE Lab and INFN entered the HBP project since its first Scientific Grant Agreement phase (SGA1, 2016-2018), guiding an international collaboration aimed at investigating the role of the different expressions of electrophysiological activity of the cortex during deep sleep. Activities went on during the SGA2 (2018-2020), followed by SGA3 in 2020 and came to a conclusion in 2023. In these years, we have consolidated our position in the field, gaining experience and extending our network and lines of actions. Our past and present research interests and efforts are focused on several items (theses available):
- hardware and software co-design for distributed spiking neural network simulations on parallel computing architectures (DPSNN)
- hardware and software co-design for neuromorphic computing systems (focus on network, latency, scalability)
- data analysis (electrophysiological and imaging data from rodents and monkeys)
- data-driven inference-based mean-field models
- thalamo-cortical spiking models of sleep and wakefulness
- software tools for the HBP-EBRAINS infrastructure platforms
ThaCo is a thalamo-cortical spiking model able to integrate some biological features of sleep states and wakefulness and perform classification tasks exploiting bio-inspired mechanisms for learning and memory formation.
Cobrawap is a Collaborative Brain Wave Analysis Pipeline developed by APE Lab together with several partners in the framework of HBP-EBRAINS.
Cobrawap collected the inheritance of the SWAP project, enhancing modularity and generalizability of the algorithms.
Further details on arXiv:2211.08527 [q-bio.NC] and GitHub
- slow waves, as the distinctive signature of the brain activity which manifests during deep sleep and constitutes our main target of research;
- multi-scales, to underline our aim of addressing the process at the different scales of investigation (microscale — the cells; mesoscale — cortical columns and cortical areas; macroscale — the bedside);
- E for Experiments, S for Simulations, since experimental data and computer-based realizations of theoretical models are the tools we leverage for our research purposes.
Simulations approaching data is the goal we aim at achieving when devising mean-field models of cortical activity. Inference methods are adopted, matched with validation techniques that allow to iteratively compare experimental target data and the outcomes of numerical simulations in order to refine model parameters and obtain cortical dynamics that contains the main features observed in the experiments reducing the amount of stereotyped signatures that generative models may contain.
DPSNN (Distributed Plastic Spiking Neural Network simulation engine) is the brain simulator developed by the APE Lab. It represents the first outcome that APE Lab delivered in the field of spiking simulation.
Originally devised as a mini-application for testing the parallel computing architectures in the framework of ExaNeSt, its performance has been improved to include short- and long-range connectivity and synaptic plasticity, and to be extended towards multi-areal cortical activity. It can accommodate different theoretical models for the neuron and the network.
Its strengths are speed, reliability and scalability, and its results are comparable to those obtained with NEST.
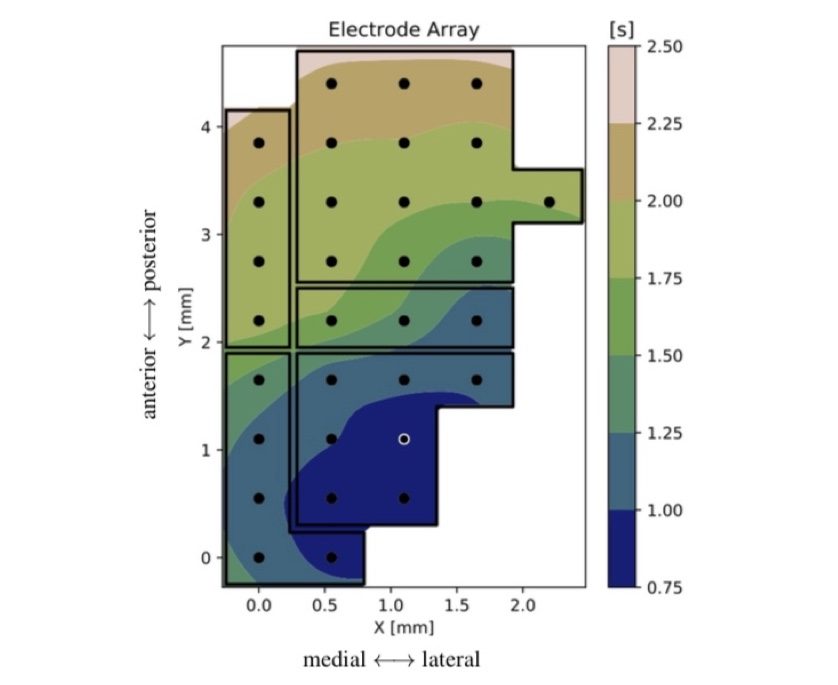
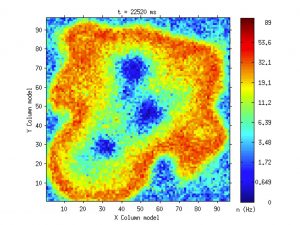
SWAP is Slow Waves Analysis Pipeline, a robust analysis procedure capable to extract, plot and statistically evaluate the features of slow oscillations and slow waves, offering a set of software tools and methods that allow going from raw-data to statistical assessment of results. It represents the first outcome that APE Lab delivered in the filed of brain data analysis.
SWAP has been largely employed in the analysis of experimental data. Currently, efforts have been directed to the improvement of the pipeline towards robustness and flexibility, aimed at extending the range of applicability (including the outcome of simulations) and at integrating it into the HBP platforms.
Among recent results obtained with SWAP, we have evaluated the differentiation of cortical areas, and we highlight the presence of gradients of excitability, in particular along a direction from fronto-lateral towards occipito-medial regions. This outcome can offer a new perspective on the classification of brain states and new input for theoretical models and data-driven simulations.
- Istituto Superiore di Sanità (ISS) (M. Mattia), Rome, Italy.
- University of Milan, Department of Clinical Sciences “L. Sacco” (M.Massimini), Milan, Italy.
- Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Sanchez-Vives Lab (M.V. Sanchez-Vives), Barcelona, Spain.
- Institute of Bioengineering of Catalonia (IBEC), Group: Nanoprobes and nanoswitches (P. Gorostiza), Barcelona, Spain.
- Università Sapienza, Department of Physiology and Pharmacology, Ferraina Lab (S. Ferraina), Rome, Italy.
- LENS, Biophysics Biophotonics Lab (F.S. Pavone), Florence, Italy
- Forschungszentrum Jülich, Institute of Neuroscience and Medicine, Computational and Systems Neuroscience (INM-6) & Theoretical Neuroscience (IAS-6), Group: Statistical Neuroscience (S. Grün), Jülich, Germany.
- Athena Research Center, Greece.
- Heidelberg University, Heidelberg, Germany, Kirchhoff-Institut für Physik, Electronic Vision(s) – BrainScales (J. Schemmel), Heidelberg, Germany.

